Madan Research Lab
Research Lab of Dr. Esha Madan
Personal Statement
Dr. Madan has expertise and research experience in the field of transcriptional biology, cell signaling and molecular oncology. At a young age, Dr. Madan made seminal discoveries in area of cancer disease regulation, cell competition and identification of genetic-networks that regulate tumor-stroma interactions and can potentially impact clinical and translational outcomes. Dr. Madan has conducted research activities across Europe, USA, and Asia, with a track record of high impact publications in journals including Nature, Trends in Cancer, Nucleic acid Research, PNAS, Cancer Research, EMBO, JBC, and MCB. Importantly, she has established a strong network of collaborations with leaders in the field, to enable utilize the state-of-art technologies, for supporting her research. Dr. Madan’s objectives are directed towards a ground-breaking research area of identifying transcriptional signals and axis in combination with epigenetics, discovery of biomarkers, and effective therapeutic intervention for the benefit of cancer patients.
During the early years of the PhD tenure (2008), Dr. Madan made important discoveries and findings in identifying the mechanisms that regulate p53-dependent apoptosis and metabolism in cancer. She published 12 first author papers during her PhD term in high impact journals and trained herself in a variety of molecular biology, biochemistry, cell culture and tumor xenografting techniques. Her collaboration with OSU during PhD gave her an opportunity to work at the interface of cancer and cardiac disease models including studying the novel role of oxygen-p53 survival pathway in cardio-protection. Dr. Madan's post-doctoral program in Switzerland (2013), trained her in molecular biology, genomics, transcriptomics, and the basics of tumor host microenvironment interactions based on 'Flower' fitness fingerprint mark in cell competition pathway, which recently got published in Nature Journal in 2019. Her research efforts were responsible for the first ever discovery of human fitness fingerprint molecules in the form of Flower Win and Lose protein.
As an independent Visiting Faculty in Portugal, Dr. Madan’s interests have been engaged in identifying the key mechanisms that regulate the competition-based interactions between the tumor and the microenvironment. She recently joined as an Assistant Professor in the Department of Surgery, VCU, where her team is uniquely poised for advancing the basic science research to clinics by effective usage of cancer disease models and human patient samples, instrumental in promoting immediate translational impact of the research findings.
Research Goals
- To understand the complex competitive interactions between a cancer cell and its microenvironment; how a cancer cell gains competitive advantage over its microenvironment. The molecular mechanisms of tumor-host interactions, cell fitness signaling and developing therapies as well as biomarkers for translational biomedical research.
- Training and education of students & researchers in the field of cell fitness and tumor microenvironment, with strong focus on basic as well as translational biomedical research. We aim to cultivate next generation scientists who have the skills to pursue successful research careers.
- Collaborating with other researchers to foster interdisciplinary research, share knowledge to a wider scientific community. We aspire to combine expertise from various fields to tackle the complex problems of biological research and gain a comprehensive understanding.
Research Focus
The ability of our body to sense and replace damaged or malfunctioning cells is fundamental in preventing infections, ageing, cardiovascular diseases, and cancers, among others. Several cellular fitness sensing mechanisms have been identified by our lab and others that play a key role in this process. The overarching goal of the lab is to identify such fitness fingerprints, understand their signaling mechanisms and utilize this knowledge in the development of therapies as well as use these as biomarkers in diseases. To tackle these, our lab utilizes molecular, biochemical and genetic tools in cells and primary human samples, as well as new cutting-edge technologies of spatial biology, single cell transcriptomics and 3-D printing. These approaches have resulted in the identification of tumor-promoting mechanisms via cell-competition in breast and ovarian cancer, biomarkers that predict COVID-19 severity, exosome-mediated long-range competitive interactions and others.
- Identification and characterization of fitness fingerprints in human diseases
Identification and characterization of fitness fingerprints in human diseases aims to understand the genetic and molecular factors contributing to disease susceptibility, progression, and response to treatments. Fitness fingerprints refer to cellular factors, such as transmembrane proteins, using which neighboring cells can sense each other’s relative health. Our lab has identified one such fitness fingerprint called Flower, using which cells can sense their relative health and participate in the phenomena called cell-competition where genetically distinct populations of cells compete for survival and resources. Studies have shown that the Flower gene is involved in regulating cell death and growth control during cell competition.
To characterize fitness fingerprints, we analyze the functional consequences of genetic and molecular alterations on cellular processes, signaling pathways, and biological networks. Moreover, large-scale data integration and bioinformatics analysis are employed to fully understand the signaling aspects of fitness fingerprints.
Fitness fingerprints may have prognostic value, helping predict disease progression, recurrence, or response to specific interventions. This information can guide clinical decision-making, facilitate early intervention, and enable timely adjustments to treatment plans.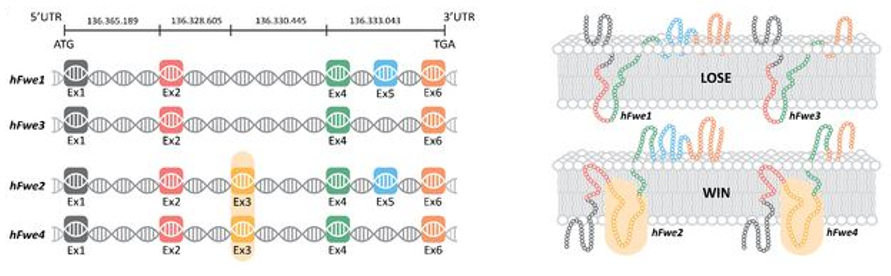
- Impact of racial diversity on cell-competition mediated disease mechanisms. There is growing recognition that genetic variations can differ among racial and ethnic groups and can have differing impact on treatment outcome. Our lab strives to understand how such differences can be encompassed within the focus of our research on fitness-fingerprints and cell competition. Towards this, we ensure that our studies such as on primary human samples are conducted in a manner representing a wide array of racial and demographic backgrounds. These variations can influence drug metabolism, treatment response, and disease susceptibility. However, it is crucial to approach race and genetics cautiously, as race is a social construct and genetic diversity exists within racial groups. The challenge lies in developing a nuanced understanding of genetic variation while considering the social and environmental factors that contribute to health disparities. Ethical considerations and equitable access to such approaches must be central to its implementation to ensure fair and effective healthcare for all individuals, irrespective of their race or ethnicity, which we believe starts at the level of basic science research and is a core focus of our lab.
- Development of Flower-targeting monoclonal antibody therapy in tumor-stroma interaction
Antibody therapy targeting cell competition in cancer is an exciting and emerging. Antibodies can be designed to specifically target and interfere with the signaling pathways involved in cell competition. These antibody therapies can disrupt the interactions between competing cells or block the signaling molecules responsible for providing a competitive advantage to certain cell populations. By doing so, they aim to restore a more balanced cellular composition within the tumor, inhibiting the expansion of aggressive and therapy-resistant cell clones. This approach holds promise for enhancing the effectiveness of existing cancer therapies by mitigating the impact of cell competition on tumor growth and evolution.
Towards this, our lab is developing monoclonal antibody (mAb) therapy against Flower which plays a central role in cell competition. Antibodies can be designed to selectively block the win or lose isoforms in this pathway. Inhibition of these factors can disrupt the competition between cells, leading to a more homogeneous and vulnerable tumor population. Antibody therapy targeting cell competition holds great potential as a novel therapeutic strategy in cancer treatment. By disrupting the competitive advantage of aggressive cell populations, these therapies aim to improve treatment outcomes, overcome therapy resistance, and pave the way for more effective and personalized cancer therapies.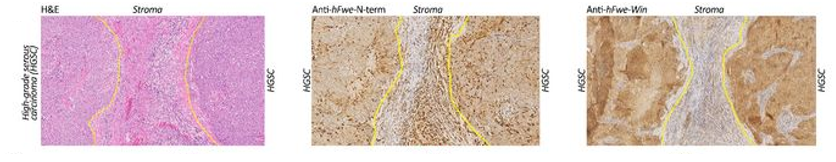
- Biomarker discovery in cancer and COVID-19 based on cellular fitness.
Biomarker discovery based on cell competition is an emerging field with the potential to revolutionize diagnosis, prognosis, and treatment of a wide variety of diseases. Cell competition creates a selective pressure that drives the emergence of fitter cells (or more aggressive cells such as in cancer). Towards this, our lab has established Flower-lose as a biomarker for COVID-19 pathogenesis. We demonstrated that Flower-lose can accurately predict the severity and hospitalization of patients. We believe that SARS-CoV2 preferentially eliminates lung alveolar cells expressing Flower-lose.
In tumors, biomarkers associated with cell competition can provide valuable information about tumor heterogeneity, aggressiveness, and therapy response. They can aid in identifying patients who are more likely to develop aggressive tumors or exhibit therapy resistance, allowing for personalized treatment strategies. Additionally, biomarkers associated with cell competition may help monitor treatment response, detect minimal residual disease, and guide therapeutic interventions. Our lab has identified soluble factors in exosomes that are directly related to tumor and stroma interactions in cancers such as ovarian cancer. We are currently developing cell-competition based fitness biomarkers that can accurately predict disease outcome, therapy resistance and may even be used for early diagnosis of a variety of different cancers through liquid biopsies.
As discussed, our lab employs technologies such as genomics, transcriptomics, and proteomics play a crucial role towards the stated biomarker discovery-goals. By analyzing the genetic and molecular alterations in cells undergoing competition, our lab aims to identify candidate biomarkers that are differentially expressed or regulated. Advanced computational and bioinformatics tools are used to mine large datasets and identify potential biomarkers that can be validated and translated into clinical practice.
Mentor Statement
At our lab, mentoring and training is core to our principles and values. We strongly believe in pushing the boundaries of existing knowledge and being comfortable with the uncharted territories of research. Our focus is on translational research from bench side to bedside. In the lab, we train the students for a wholistic approach to research, including hypothesis testing, being innovative in their approach to problem solving, data analysis, improving presentation & communication skills as well as training them to write grants for fund raising. We strongly believe in collaborations and interdisciplinary research. The trainees are encouraged to participate in departmental and institutional meetings where they would present their work. In addition, students are encouraged to present their findings in conferences through oral and/ or poster presentations. A cornerstone of our lab is to publish our scientific findings in academic journals for dissemination of such knowledge to the wider scientific community. I have trained several undergraduates, medical students, PhD and MS students and post-doctoral fellows at the lab.
Lab Members
Dr. Antonio Palma, Ph.D.

My research focus has been in the field of cancer biology with a particular interest in transcriptional mechanisms, cell signaling, and cell competition. My prior training includes a Bachelor's in Medical Laboratory Sciences (2015), a Master's in Oncobiology (2019), and a PhD in Biotechnology and Biosciences (2023). Since 2016, my main goal is to study cancer biology from a translational perspective, which aims to bring basic science research findings from the bench to clinics. This can be achieved by using human patient samples to describe cancer models, which can later be used to discover new early detection and prognosis biomarkers and to find new therapeutic targets that will improve cancer treatment. In 2019, I joined Dr Madan and Dr Gogna's team and collaborated on the publications at multiple high-impact journals such as Nature, PNAS, NAR, Cancer Research, EMBO Mol Med, and EMBO Journal. Among th
em, I highlight the discovery of fitness fingerprints and cell competition mechanisms in the promotion of tumor growth, published in Nature (2019), which regulatory mechanisms were further elucidated during my PhD training. My scientific contributions are not limited to cancer biology. Together with Dr Madan, I published a study that discovered Flower Lose as a prognostic marker for COVID-19 outcomes. Additionally, I have a publication on embryonic development that applies artificial intelligence and machine learning tools to describe how cell-cell interactions affect the transcriptome in a cell-type-specific manner. In the future, I intend to continue studying cell competition and cellular microenvironments in different diseases, thus finding potential biomarkers and therapeutic targets that will improve patients' health and secure continuous publications in high-impact journals.
- Palma AM, Bushnell GG, Wicha MS, Gogna R. Tumor Microenvironment interactions with cancer stem cells in pancreatic ductal adenocarcinoma. Advances in Cancer Research. Forthcoming; 159. Available from: https://www.elsevier.com/books/pancreatic-cancer-basic-mechanisms-and-therapies/emdad/978-0-443-13354-1 First Author
- Palma AM, Peixoto MLeonor, Vudatha V, Madan E. Tumor heterogeneity: an oncogenic driver of PDAC progression and therapy resistance under stress conditions. Advances in Cancer Research. Forthcoming; 159.Available from: https://www.elsevier.com/books/pancreatic-cancer-basic-mechanisms-and-therapies/emdad/978-0-443-13354-1 First Author
- KimJ, Rothová M, Madan E, Rhee S, Weng G, Palma A, Liao L, David E, Amit I,Hajkarim M, Vudatha V, Gutiérrez-García A, Moreno E, Winn R, Trevino J, FisherP, Brickman J, Gogna R, Won K. Neighbor-specific gene expression revealed from physically interacting cells during mouse embryonic development. Proceedings oft he National Academy of Sciences. 2023 January 03; 120(2):-.Available from: https://pnas.org/doi/10.1073/pnas.2205371120 DOI:10.1073/pnas.2205371120
- Madan E, Palma AM, Vudatha V, Trevino JG, Natarajan KN,Winn RA, Won KJ, Graham TA, Drapkin R, McDonald SAC, Fisher PB, Gogna R. CellCompetition in Carcinogenesis. Cancer Res. 2022 Dec 16;82(24):4487-4496. PubMedCentral PMCID: PMC9976200. Shared First Author
- Madan E,Parker TM, Pelham CJ, Palma AM, Peixoto ML, Nagane M, Chandaria A, Tomás AR,Canas-Marques R, Henriques V, Galzerano A, Cabral-Teixeira J, Selvendiran K,Kuppusamy P, Carvalho C, Beltran A, Moreno E, Pati UK, Gogna R. HIF-transcribedp53 chaperones HIF-1α. NucleicAcids Res. 2019 Nov 4;47(19):10212-10234. PubMed Central PMCID: PMC6821315.
- Madan E, Pelham CJ, Nagane M, Parker TM, Canas-MarquesR, Fazio K, Shaik K, Yuan Y, Henriques V, Galzerano A, Yamashita T, Pinto MAF,Palma AM, Camacho D, Vieira A, Soldini D, Nakshatri H, Post SR, Rhiner C,Yamashita H, Accardi D, Hansen LA, Carvalho C, Beltran AL, Kuppusamy P, GognaR, Moreno E. Flower isoforms promote competitive growth in cancer. Nature.2019 Aug;572(7768):260-264. PubMed PMID: 31341286
Anaya Surve
 My name is Anaya Surve and I am a bioinformatics major on the pre-med track. When I am not in the lab or helping edit papers, I am volunteering at ASK Childhood Cancer Foundation, tutoring at the Campus Learning Center, or playing cards with my brother. I value the importance of research in driving medicine forward and am very grateful and excited to be part of that process
My name is Anaya Surve and I am a bioinformatics major on the pre-med track. When I am not in the lab or helping edit papers, I am volunteering at ASK Childhood Cancer Foundation, tutoring at the Campus Learning Center, or playing cards with my brother. I value the importance of research in driving medicine forward and am very grateful and excited to be part of that process
Dr. Sahil Chaudhary, M.B.B.S. – Research Assistant
 Translational medicine is the bridge that connects basic sciences research to the bedside clinical application. My interest in cellular and molecular biology, motivated me to pursue an engineering degree in Biotechnology. During my engineering, I understood the complex cell signaling pathways, and the technology of genetic engineering in curing diseases and that drew my attention towards oncology. After understanding biology at a cellular & molecular level during my undergrad, I wanted to understand the complex human body as well as their diseases and their management; this nudged me in the direction of medical school. My training as a medical doctor helped me understand not only the clinical presentation of a diseases and their management but also the importance of translational medicine in the clinical setting. During the Covid-19 pandemic, while working as a junior doctor in emergency medicine, I worked with Dr. Gogna & Dr. Madan and contributed in identifying one cell fitness marker called “Flower” in Covid-19 patients. In Dr. Madan’s lab, I will use my medical training and experience in clinical pathology to understand the basic science mechanisms of competitive cell interactions and their impact on a wide array of diseases. I will bring in translational knowledge to develop monoclonal antibody therapies to leverage the basic science mechanisms discovered in the lab. Outside the lab, I am a fitness enthusiast and excited about adventure sports. On lazy days, I like to spend my time playing open-world games on PlayStation.
Translational medicine is the bridge that connects basic sciences research to the bedside clinical application. My interest in cellular and molecular biology, motivated me to pursue an engineering degree in Biotechnology. During my engineering, I understood the complex cell signaling pathways, and the technology of genetic engineering in curing diseases and that drew my attention towards oncology. After understanding biology at a cellular & molecular level during my undergrad, I wanted to understand the complex human body as well as their diseases and their management; this nudged me in the direction of medical school. My training as a medical doctor helped me understand not only the clinical presentation of a diseases and their management but also the importance of translational medicine in the clinical setting. During the Covid-19 pandemic, while working as a junior doctor in emergency medicine, I worked with Dr. Gogna & Dr. Madan and contributed in identifying one cell fitness marker called “Flower” in Covid-19 patients. In Dr. Madan’s lab, I will use my medical training and experience in clinical pathology to understand the basic science mechanisms of competitive cell interactions and their impact on a wide array of diseases. I will bring in translational knowledge to develop monoclonal antibody therapies to leverage the basic science mechanisms discovered in the lab. Outside the lab, I am a fitness enthusiast and excited about adventure sports. On lazy days, I like to spend my time playing open-world games on PlayStation.
Affiliated Lab Members
Dr. Praveen Bhoopathi, Ph.D.
 Leveraging my background in cancer biology, I have conducted extensive research aimed at understanding the molecular basis of various types of cancer, including neuroblastoma, glioblastoma, pancreatic cancer, prostate cancer, and glioma. My research endeavors encompass a wide range of topics within cancer biology, with a particular focus on studying the mechanisms underlying cancer progression, metastasis, and the development of effective therapies.
Leveraging my background in cancer biology, I have conducted extensive research aimed at understanding the molecular basis of various types of cancer, including neuroblastoma, glioblastoma, pancreatic cancer, prostate cancer, and glioma. My research endeavors encompass a wide range of topics within cancer biology, with a particular focus on studying the mechanisms underlying cancer progression, metastasis, and the development of effective therapies.
In the realm of gene therapy, I have explored its potential application as a therapeutic approach for these different types of cancer. By investigating the interactions between genetic alterations, signaling pathways, and cellular processes, I aim to uncover new insights into the biology of these cancers and identify potential targets for therapeutic intervention.
Through my efforts, I have contributed to the scientific community with approximately 50 peer-reviewed publications published in prestigious journals such as PNAS, Cancer Research, and Cell Death and Differentiation. These publications have highlighted the significance of gene therapy in preventing metastasis, inducing tumor regression, and advancing our understanding of cancer.
By expanding our knowledge of the molecular mechanisms underlying these cancers, I strive to contribute to the development of innovative and effective treatment strategies that can ultimately improve patient outcomes in the field of cancer biology.
Dr. Amit Kumar, Ph.D.
My academic training and research experience have provided me with an excellent background in multiple biological disciplines including molecular biology, microbiology, cell biology, and microscopy. In my early career I studied host-pathogen interactions and molecular pathogenesis, intracellular signaling, and cytotoxicity. In my work, during graduate and as a postdoctoral fellow, I have applied traditional and innovative cell biology, transcriptomic and flow cytometric approaches to study KSHV reactivation and EBV mechanisms of genomic instability in nasopharyngeal carcinoma and got expertise in live cell imaging and genomic instability. The postdoctoral research at JHMI provided me with new conceptual and technical training in stem cell biology and genome editing. I had been conducting research to develop a hiPSC- derived HSPC-based therapy as a curative alternative to the expensive non-curative GCase enzyme replacement therapy (ERT) to treat Gaucher's Disease (GD). Parallelly at Johns Hopkins I explored and understand the complexities of cancer immunotherapy and cell-cell interactions, I used immunotoxin (DAB-IL2) directed to deplete Tregs from the tumor microenvironment in different established preclinical tumor models and found that Tregs depletion from tumor microenvironment resulted in the antitumor effect and enhanced immune functions. Currently I am working as a postdoctoral fellow at VCU to define the molecular basis of cancer development and progression to metastasis. In the future, I am planning to continue research on the interactions between cells and the cellular microenvironments in various diseases in order to identify possible biomarkers and therapeutic targets that will enhance patient health.
Anza Sardar
 As a Biology major with a minor in Political Science at VCU, I am dedicated to pursuing a Bachelors in the pre-medical track followed by a degree as Doctor in Medicine. Throughout my introductory and advanced coursework, my passion for science has flourished, driving me to seek independent research opportunities as an undergraduate student. My specific interests lie in molecular and cellular biology as well as genetics. My recent research focus has been on the correlation between dopamine activity ratio, measured by radiolabella (L-dopa) and positron emission tomography (PET), and the potential increase in presynaptic striatal dopaminergic function in schizophrenic patients. By exploring the influence of risk factors such as trauma, drug use, and genetics, I aim to shed light on this complex relationship. Currently (2023), I had the privilege of joining the esteemed team led by Dr. Gogna and Dr. Madan. As part of this collaborative effort, we embark on groundbreaking research projects that result in the publication of our work in multiple high-impact journals. Moving forward, my academic and research pursuits will be focused on studying the intricate topics of molecular and cellular biology, with a specific emphasis on genetics as it relates to various diseases. I aspire to uncover key molecular pathways, genetic variations, and bimolecular signatures that can serve as diagnostic tools and guide the development of targeted therapies that have the potential to enhance patients' health and well-being.
As a Biology major with a minor in Political Science at VCU, I am dedicated to pursuing a Bachelors in the pre-medical track followed by a degree as Doctor in Medicine. Throughout my introductory and advanced coursework, my passion for science has flourished, driving me to seek independent research opportunities as an undergraduate student. My specific interests lie in molecular and cellular biology as well as genetics. My recent research focus has been on the correlation between dopamine activity ratio, measured by radiolabella (L-dopa) and positron emission tomography (PET), and the potential increase in presynaptic striatal dopaminergic function in schizophrenic patients. By exploring the influence of risk factors such as trauma, drug use, and genetics, I aim to shed light on this complex relationship. Currently (2023), I had the privilege of joining the esteemed team led by Dr. Gogna and Dr. Madan. As part of this collaborative effort, we embark on groundbreaking research projects that result in the publication of our work in multiple high-impact journals. Moving forward, my academic and research pursuits will be focused on studying the intricate topics of molecular and cellular biology, with a specific emphasis on genetics as it relates to various diseases. I aspire to uncover key molecular pathways, genetic variations, and bimolecular signatures that can serve as diagnostic tools and guide the development of targeted therapies that have the potential to enhance patients' health and well-being.
Dr. Kartik Gupta, Ph.D
 I obtained my Ph.D. in cell and molecular biology at the University of Wisconsin-- Madison where I investigated the role of cell death in cancer and cardiovascular biology. I investigated the role of post-translational modifications on key cell death proteins in the regulation of cell-death and identified a novel pathway of exosome secretion in the context of cell death. I have been a part of Dr. Esha Madan and Dr. Rajan Gogna's group for 10+ years and have contributed to several discoveries in their team related to cell competition and oxygen-signaling. After my Ph.D., I was a post-doc at Merck and Co., and am currently a scientist in immuno-oncology at a Bay Area startup. In my free time I am passionate about weightlifting, running, and watching TV shows.
I obtained my Ph.D. in cell and molecular biology at the University of Wisconsin-- Madison where I investigated the role of cell death in cancer and cardiovascular biology. I investigated the role of post-translational modifications on key cell death proteins in the regulation of cell-death and identified a novel pathway of exosome secretion in the context of cell death. I have been a part of Dr. Esha Madan and Dr. Rajan Gogna's group for 10+ years and have contributed to several discoveries in their team related to cell competition and oxygen-signaling. After my Ph.D., I was a post-doc at Merck and Co., and am currently a scientist in immuno-oncology at a Bay Area startup. In my free time I am passionate about weightlifting, running, and watching TV shows.
Maria Leonor Peixoto
 In 2019 I was fortunate enough to be committed to research work concerning tumors and their microenvironment- I have studied the alternative splicing that generates the Win and Lose Flower isoforms (Madan E. et al., Nature 2019). I eventually completed the integrated MSc in Biological Engineering from the Instituto Superior Tecnico (Lisbon) and continued my scientific career as a Ph.D. Student of Biotechnology and Biosciences. My training and expertise achieved during the last four years under the supervision of Dr. Esha Madan are in the field of molecular oncology and deal with the role of transcription factors in cancer growth and metastasis. My work was translated into publications and reviews in high-impact journals such as NAR and Trends Cell Biology. I aim to continue performing cutting-edge research and bring innovative and newly identified biomarkers for the early diagnosis and development of anti-cancer therapies from the bench to the bedside.
In 2019 I was fortunate enough to be committed to research work concerning tumors and their microenvironment- I have studied the alternative splicing that generates the Win and Lose Flower isoforms (Madan E. et al., Nature 2019). I eventually completed the integrated MSc in Biological Engineering from the Instituto Superior Tecnico (Lisbon) and continued my scientific career as a Ph.D. Student of Biotechnology and Biosciences. My training and expertise achieved during the last four years under the supervision of Dr. Esha Madan are in the field of molecular oncology and deal with the role of transcription factors in cancer growth and metastasis. My work was translated into publications and reviews in high-impact journals such as NAR and Trends Cell Biology. I aim to continue performing cutting-edge research and bring innovative and newly identified biomarkers for the early diagnosis and development of anti-cancer therapies from the bench to the bedside.
- Palma AM, Peixoto Maria Leonor, Vudatha V, Madan E. Tumor heterogeneity: an oncogenic driver of PDAC progression and therapy resistance under stress conditions. Advances in Cancer Research, 159 (2023);
- Esha Madan, Maria Leonor Peixoto, Peter Dimitrion, Timothy D. Eubank, Michail Yekelchyk, Sarmistha Talukdar, Paul B. Fisher, Qing-Sheng Mi, Eduardo Moreno, Rajan Gogna Cell Competition Boosts Clonal Evolution and Hypoxic Selection in Cancer. Trends in Cell Biology, 30, 12, 967-978 (2020);
- Camacho D, Jesus JP, Palma AM, Martins SA, Afonso A, Peixoto Maria Leonor, Pelham CJ, Moreno E, Gogna R., SPARC-p53: The double agents of cancer. Advances in Cancer Research, 148:171-199 (2020);
- Pelham C, Parker TM, Nagane M, Beltran A, Carvalho C, Selvendiran K, Pati U, Maria Leonor Peixoto, Gogna R., HIF-transcribed p53 chaperones HIF-1α in an oncogenic feed-back loop. Nucleic Acid Research, 47, 19, 10212–10234 (2019).
Selected Peer-Reviewed Publications
1. Kim J, Rothová MM, Madan E, Rhee S, Weng G, Palma AM, Liao L, David E, Amit I, Hajkarim MC, Vudatha V, Gutiérrez-García A, Moreno E, Winn R, Trevino J, Fisher PB, Brickman JM, Gogna R, Won KJ. Neighbor-specific gene expression revealed from physically interacting cells during mouse embryonic development. Proc Natl Acad Sci U S A. 2023 Jan 10;120(2):e2205371120. doi: 10.1073/pnas.2205371120. Epub 2023 Jan 3. PMID: 36595695; PMCID: PMC9926237. https://doi.org/10.1073/pnas.2205371120
2. Madan E, Pelham CJ, Nagane M, Parker TM, Canas-Marques R, Fazio K, Shaik K, Yuan Y, Henriques V, Galzerano A, Yamashita T, Pinto MAF, Palma AM, Camacho D, Vieira A, Soldini D, Nakshatri H, Post SR, Rhiner C, Yamashita H, Accardi D, Hansen LA, Carvalho C, Beltran AL, Kuppusamy P, Gogna R, Moreno E. Flower isoforms promote competitive growth in cancer. Nature. 2019 Aug;572(7768):260-264. doi: 10.1038/s41586-019-1429-3. Epub 2019 Jul 24. PubMed PMID: 31341286. https://www.nature.com/articles/s41586-019-1429-3
3. Madan E, Palma AM, Vudatha V, Trevino JG, Natarajan KN, Winn RA, Won KJ, Graham TA, Drapkin R, McDonald SAC, Fisher PB, Gogna R. Cell Competition in Carcinogenesis. Cancer Res. 2022 Dec 16;82(24):4487-4496. doi: 10.1158/0008-5472.CAN-22-2217. PMID: 36214625; PMCID: PMC9976200. https://doi.org/10.1158/0008-5472.CAN-22-2217
4. Yekelchyk M, Madan E, Wilhelm J, Short KR, Palma AM, Liao L, Camacho D, Nkadori E, Winters MT, Rice ES, Rolim I, Cruz-Duarte R, Pelham CJ, Nagane M, Gupta K, Chaudhary S, Braun T, Pillappa R, Parker MS, Menter T, Matter M, Haslbauer JD, Tolnay M, Galior KD, Matkwoskyj KA, McGregor SM, Muller LK, Rakha EA, Lopez-Beltran A, Drapkin R, Ackermann M, Fisher PB, Grossman SR, Godwin AK, Kulasinghe A, Martinez I, Marsh CB, Tang B, Wicha MS, Won KJ, Tzankov A, Moreno E, Gogna R. Flower lose, a cell fitness marker, predicts COVID-19 prognosis. EMBO Mol Med. 2021 Nov 8;13(11):e13714. doi: 10.15252/emmm.202013714. Epub 2021 Oct 18. PMID: 34661368; PMCID: PMC8573598. https://www.embopress.org/doi/full/10.15252/emmm.202013714
5. Parker TM, Gupta K, Palma AM, Yekelchyk M, Fisher PB, Grossman SR, Won KJ, Madan E, Moreno E, Gogna R. Cell competition in intratumoral and tumor microenvironment interactions. EMBO J. 2021 Sep 1;40(17):e107271. doi: 10.15252/embj.2020107271. Epub 2021 Aug 9. PMID: 34368984; PMCID: PMC8408594. https://doi.org/10.15252/embj.2020107271
Complete list of published work in My Bibliography: https://www.ncbi.nlm.nih.gov/myncbi/1pc9Ct9N0rzQh/bibliography/public/
